
Molecular Physiology
About us
We are intrigued by how we can utilize the setting of normal embryonic development to understand tumor initiation. We have a highly transdisciplinary approach, merging the worlds of developmental and cancer biology.
We focus on the childhood tumor form neuroblastoma and adult tumor form paraganglioma. These cancers originate from cells of the sympathetic nervous system (SNS), in turn derived from the transient embryonic neural crest stem cell population. Neuroblastoma is the most common tumor form in infants. It is believed to arise due to developmental defects during embryogenesis, but the precise origin and cause of tumor is unknown. Paragangliomas are on the other hand slow growing tumors and mostly benign, but also these are believed to be primed during development of the neural crest. Neuroblastoma is the deadliest cancer in children, and for the 10% of paraganglioma patients with metastases, there are no treatment options.
We aim to decipher the mechanisms of tumor initiation during embryogenesis and thereby find diagnostic and prognostic biomarkers and improved treatment options.
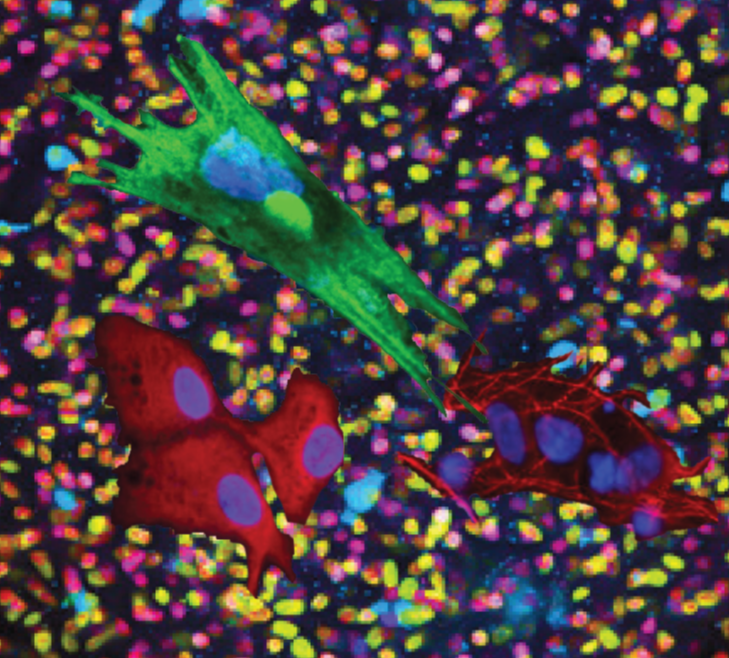
Maintaining multipotent trunk neural crest stem cells as self-renewing crestospheres, Dev Biol 2019
Research
Our main project lines:
- Establish a human-chick chimera disease model
- Investigate the biology behind HIF-2a-driven disease
- Map gene regulatory networks connecting embryonic development and tumor initiation
- Investigate the micro-environmental regulation of cancer cell fate plasticity
Models and organisms
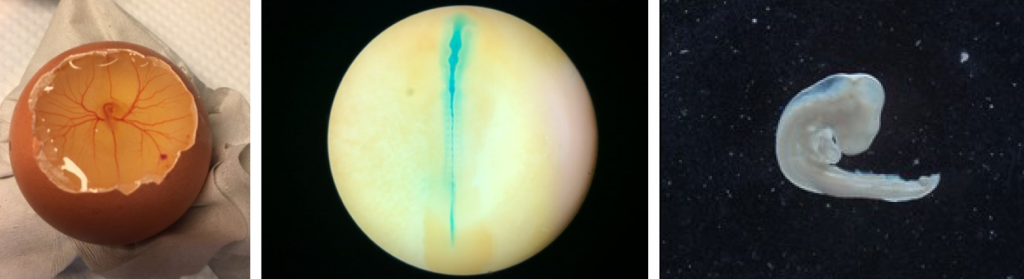
Our main model organism is the chick embryo, ideal for functional and translational studies on neural crest development. Chick embryos are easily accessible, and their nervous system development is similar to that of human embryos at comparable stages. To complement the in vivo system of chick embryos, we employ patient omics data, single cell RNA sequencing, human and mouse embryos, human cancer cell cultures, human pluripotent stem cells, chick embryo neural crest-derived in vitro crestosphere cultures, and zebrafish and mouse in vivo models.
Methodology
We always aim the use the most appropriate methodology to answer each specific research question independent of whether it is within or outside our comfort zone. Methods and material that we use include, but are not limited to:
- Human, Mouse and Chick embryos
- Human cell cultures
- Zebrafish models
- In vivo mouse xenograft models
- Bulk- and single cell RNA sequencing
- In ovo electroporations
- CRISPR/Cas9
- Conditional cell knockout in vivo models
- Chick embryo derived in vitro crestosphere stem cell cultures
- Neural tube explants
- In vivo real-time imaging

